Flemme: A FLExible and Modular Learning Platform for MEdical Images
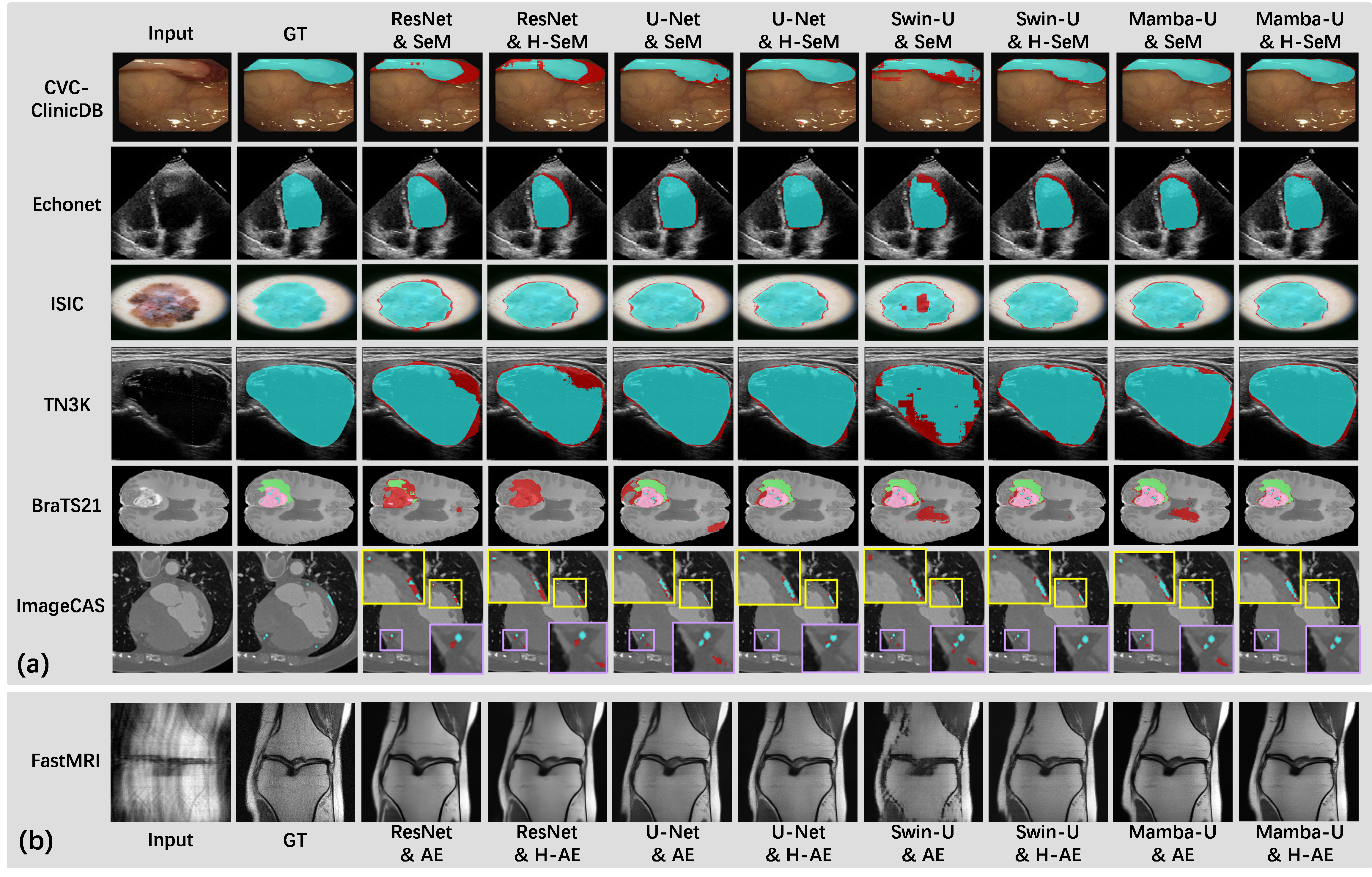
Examples of using FLEMME for testing backbone and decoder architectures on downstream medical image understanding tasks.
Motivation
Since AlexNet competed in the ImageNet Large Scale Visual Recognition Challenge, convolutional neural networks (CNNs) have become dominant in computer vision. In particular, the most preferred choice in medical imaging is U-Net which uses skip connections to enhance feature fusion across different time stages. In recent years, treating images as sequences has garnered significant attention. New models based on Vision Transformer and Vision Mamba have consistently achieved state-of-the-art results in medical image segmentation across multiple datasets.
Although new methods continue to emerge, several aspects remain worthy of exploration. Firstly, model performance depends highly on actual training techniques and deployment. In real-world applications, the improvement in model performance needs to be balanced against the computational costs of training and inference. Secondly, beyond classification and segmentation, powerful model architectures have been proposed for image reconstruction and generation, such as variational auto-encoder (VAE) and diffusion models. Applying the advanced backbones to these architectures for medical images is promising and worth anticipating. Thirdly, medical images often lack large-scale, high-quality annotations but encompass a wide range of modalities, making it hard to train a general model that performs well across various medical image datasets. In practice, researchers and engineers often need to manually build and test models with multiple backbones and architectures for specific tasks. However, combining independent methods can be labor-intensive and hard to analyze.
Given the aforementioned challenges, the lack of a universal platform that supports the fast construction of diverse models significantly adds barriers to adopting the latest technologies and increases research duration. We propose Flemme, a FLExible and Modular learning platform for MEdical images, which is distinguished by the following features:
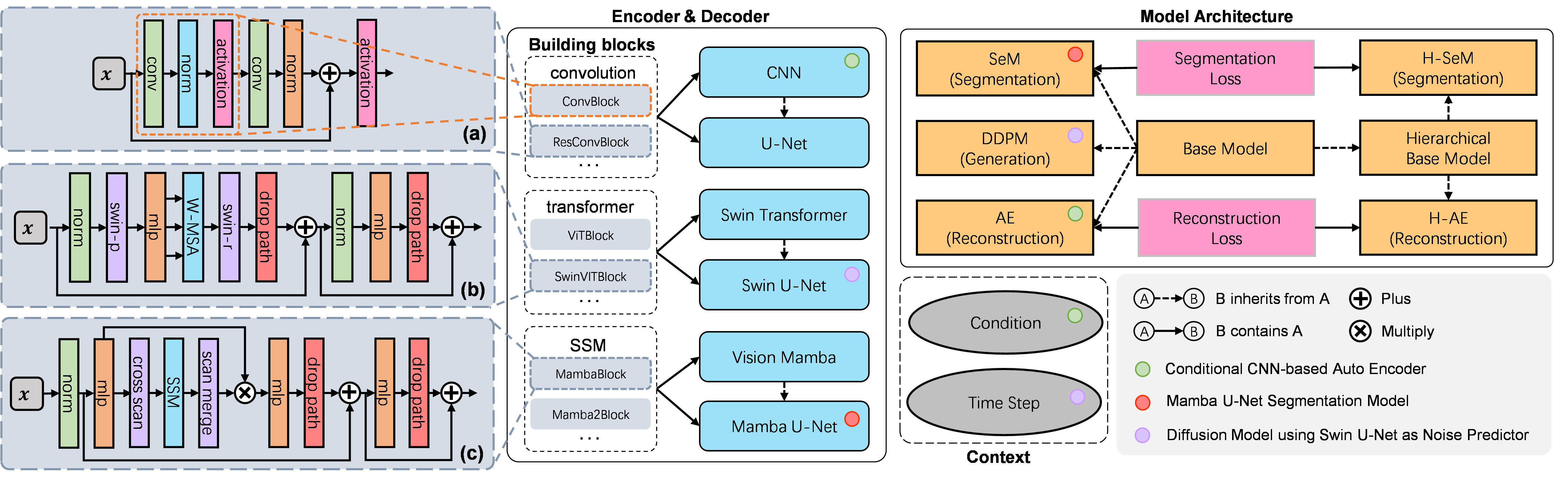
We adopt a modular design with state-of-the-art encoders and architectures for fast model construction.
We implement a novel backbone-agnostic hierarchical architecture combining a pyramid loss for generic vertical feature fusion and optimization.
We support flexible context encoding and allow extensions of new modules for more data types and tasks.
Overview of the FLEMME library
We demonstrate the applications of Flemme on various medical image datasets. For all supported models, we can set hyper-parameters such as network depth, normalization, and training strategies in the same manner. Benefiting from this advantage, we conduct extensive experiments with various encoders on different tasks to fairly compare and analyze the performance, as well as the time and memory complexity. We hope our work will set new benchmarks and serve as a valuable research tool for future investigations into the potential of convolutions, transformers, and SSMs in medical imaging.
Method
Model Architecture
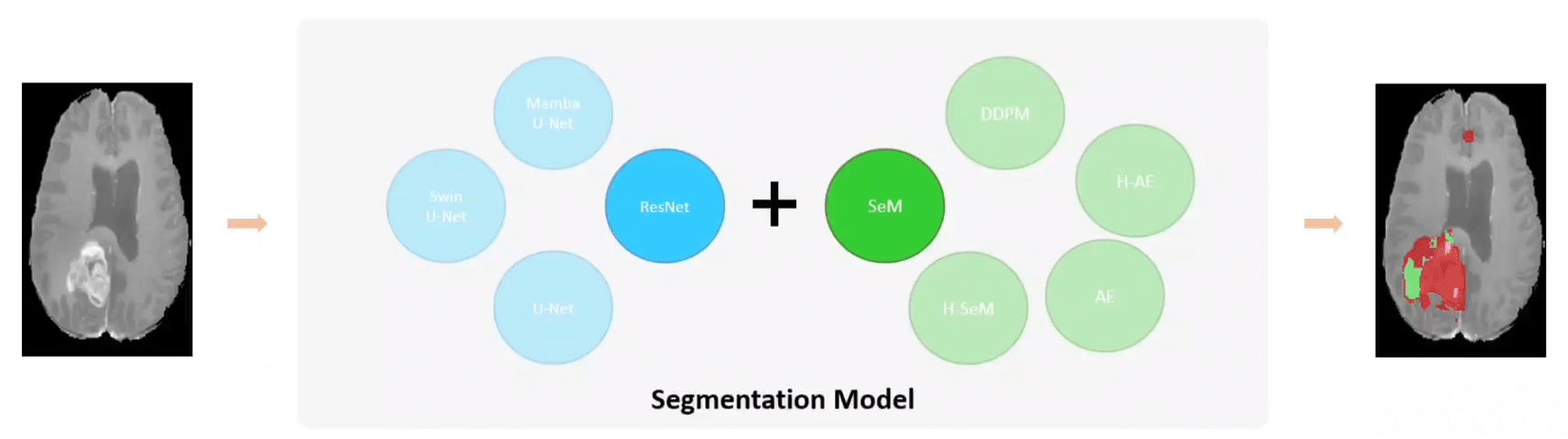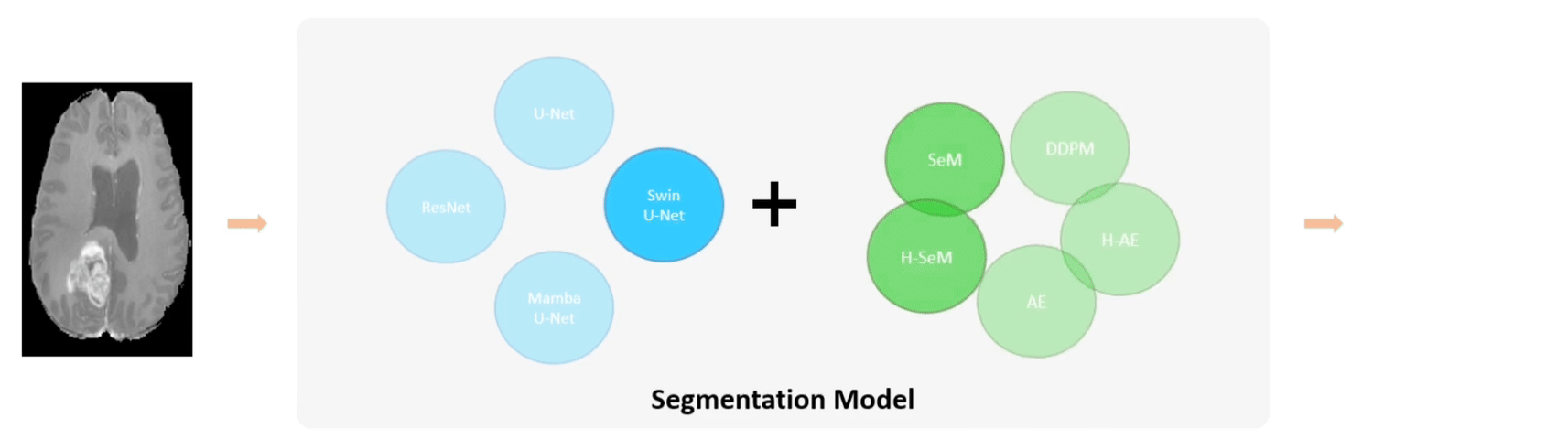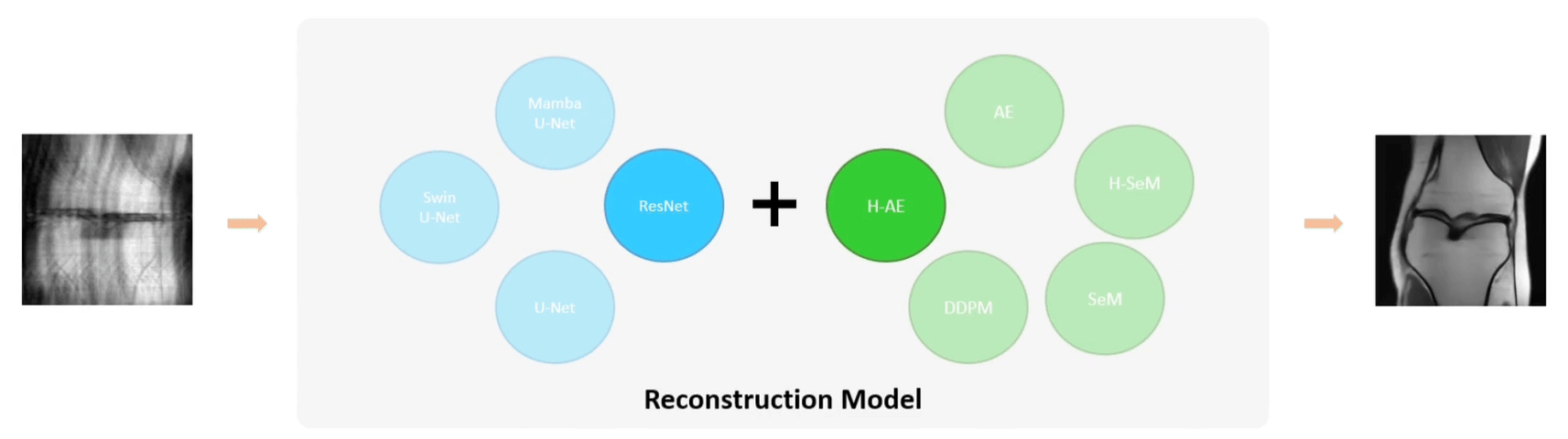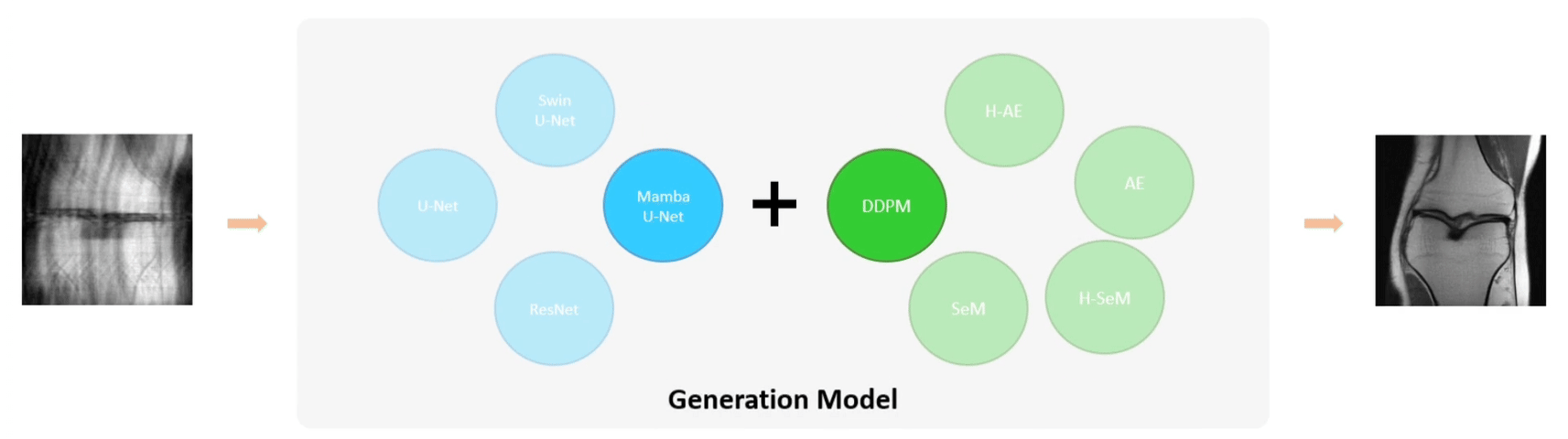
The base model architecture consists of an encoder, a decoder, and their corresponding conditional encodings. We support different types of conditions such as class labels, images, position or time step indices. We further extend the base model architecture to SeM (Segmentation Model), AE (Auto-Encoder), and DDPM (Denoising Diffusion Probabilistic Model) for medical image segmentation, reconstruction, and generation, respectively.
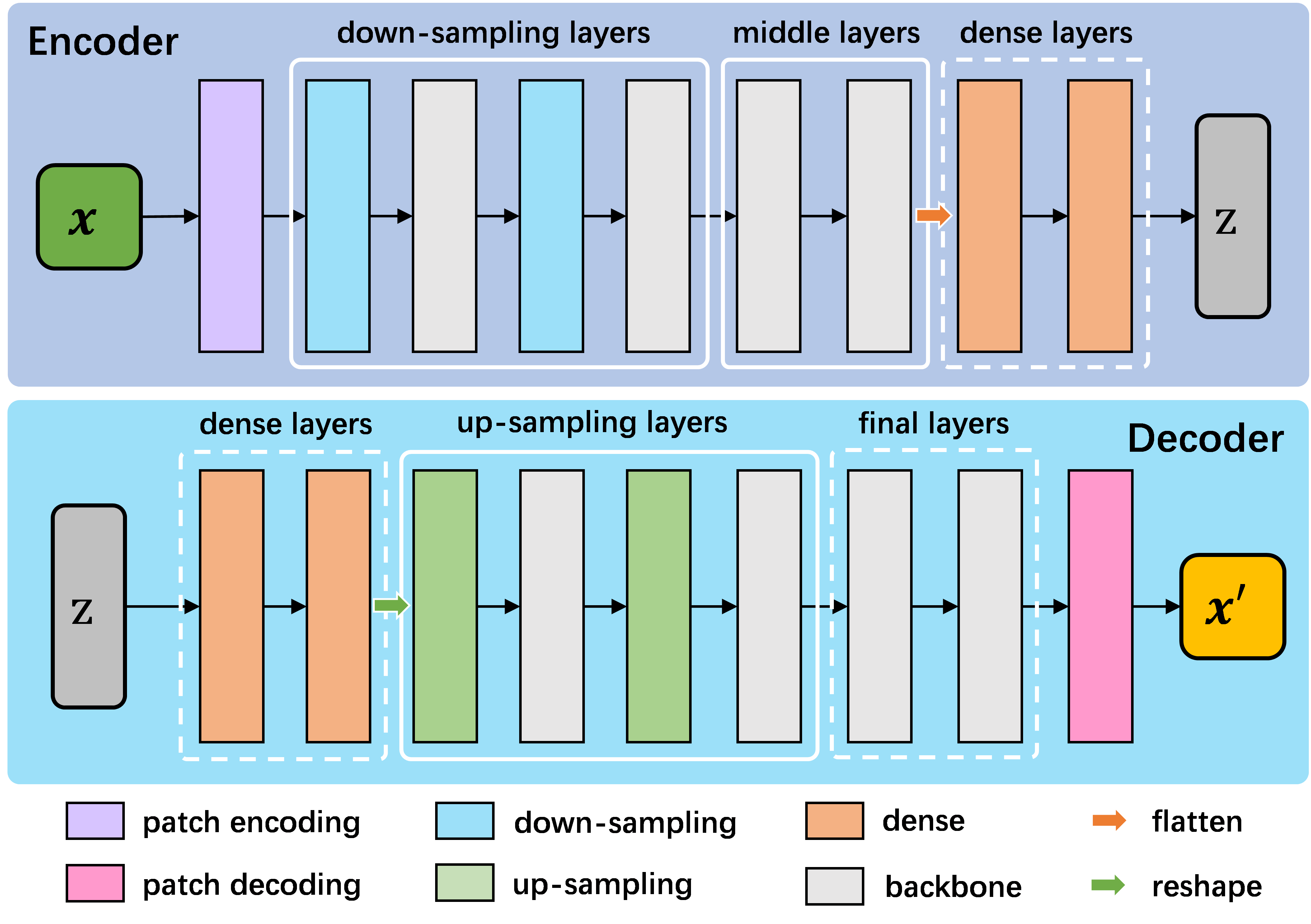
Encoder and Decoder
As shown in the above figure, our encoder first encodes the image into image patches, which are then processed through downsampling layers, middle layers, and optional dense layers to extract latent features. The decoder reverses this process to reconstruct the target-space image from the latent features. The primary distinction between different encoders lies in their building blocks. We support multiple building blocks based on CNN, Vision Transformer, and Vision Mamba.
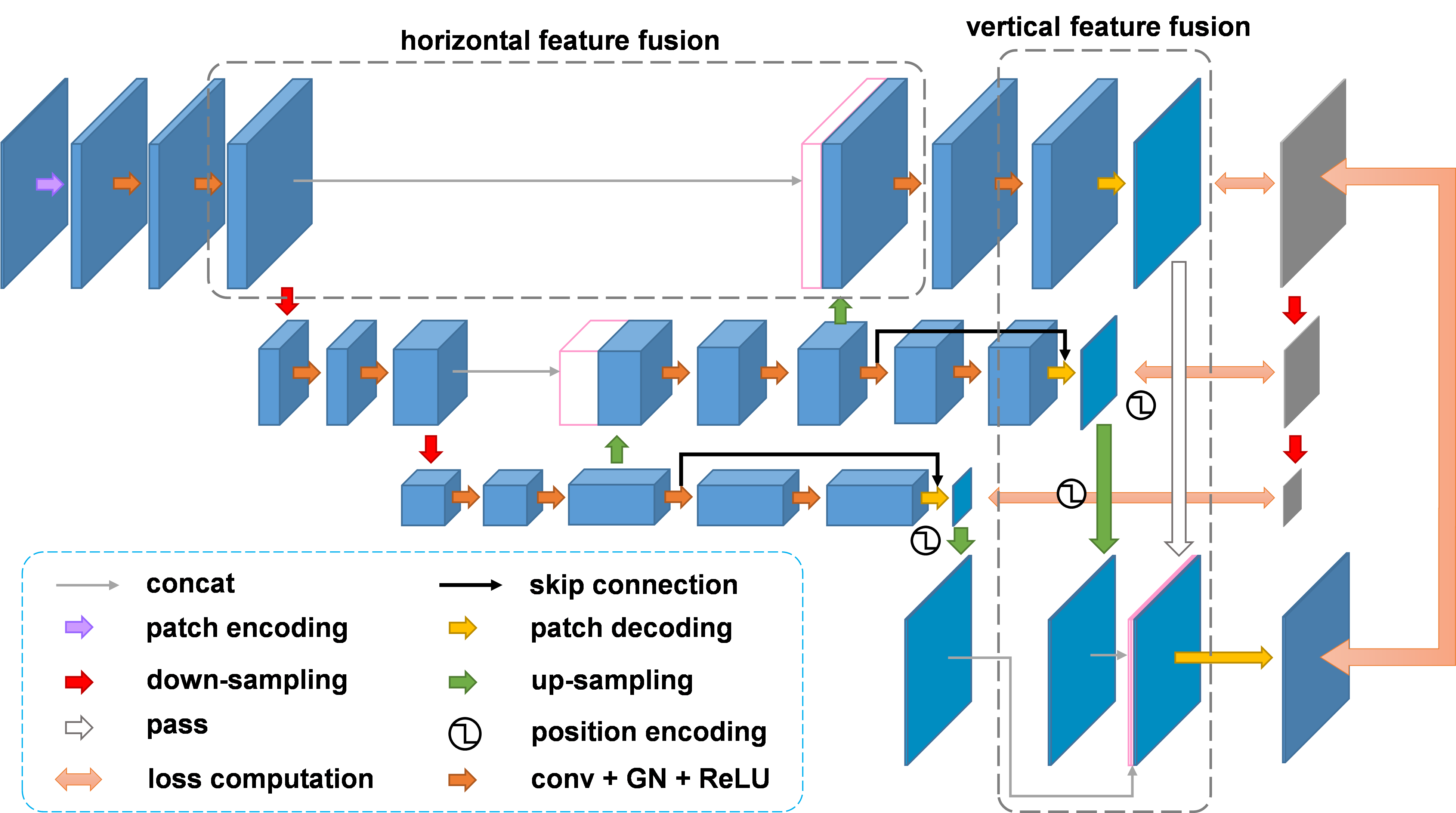
U-Shaped Networks and Hierarchical Architecture
Several studies have demonstrated the superiority and robustness of U-shaped networks in image segmentation and reconstruction. Therefore, we created U-shaped counterparts for all supported encoders. The U-shaped design essentially serve as a horizontal feature fusion mechanism. In addition, we propose a general multi-scale vertical feature fusion architecture, combined with pyramid-based loss functions, to enhance model performance in segmentation and reconstruction tasks. The hierarchical versions of SeM and AE are denoted as H-SeM and H-AE, respectively.
Results
We conduct extensive experiments to compare and analyze the performance differences between various encoder-decoder architectures using Flemme. Our proposed vertical feature fusion architecture consistently improves the performance over different encoders in both reconstruction and segmentation tasks. From the encoder's point of view, Mamba-based methods show the highest superiority. U-Net also demonstrates satisfactory performance, achieving comparable performance to Swin Transformer in reconstruction accuracy and outperforming it in segmentation accuracy. We notice that sequence modeling-based encoders generally outperform convolution-based encoders for reconstruction and generation, indicating that exploring long-range dependency is particularly effective for high-frequency component prediction.
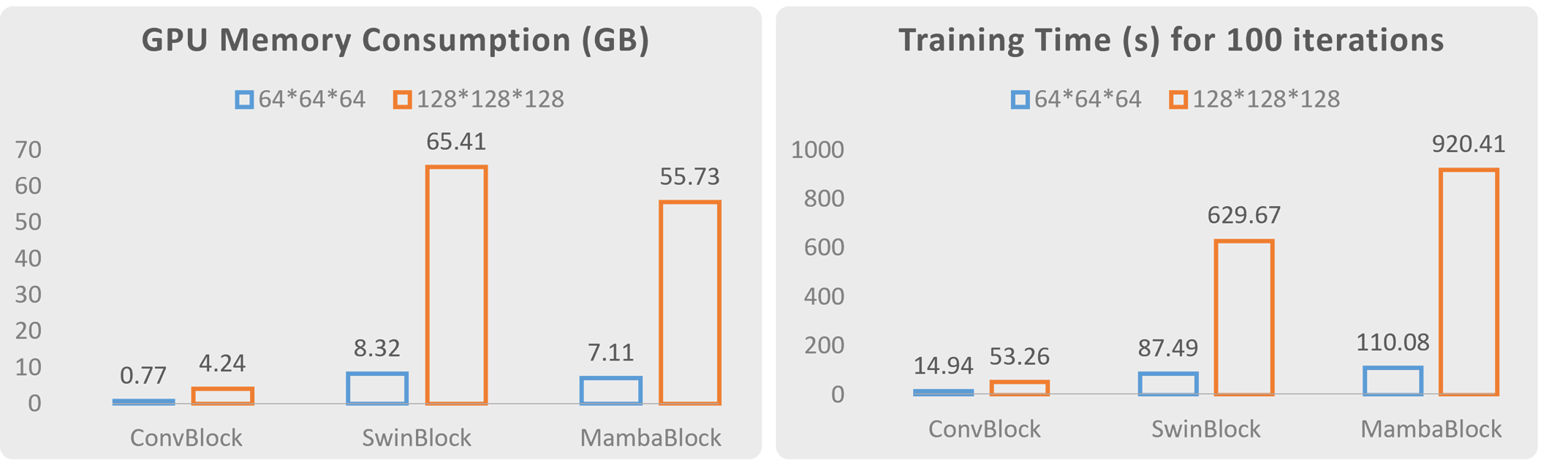
We also compare the training time and GPU memory consumption of different building blocks. As the size of 3D image patches continues to increase, the advantages of CNNs in terms of training and inference costs become significantly more pronounced compared to Vision Transformer and Vision Mamba. Balancing training and inference costs with model performance, we believe that CNNs will continue to hold an irreplaceable position in medical image segmentation and reconstruction.
Conclusion
We present Flemme, a general learning platform aiming at the rapid and flexible development of deep learning models for medical images. We introduce various encoders based on convolution, transformer, and SSM backbones, combining SeM, AE, and DDPM architectures to facilitate model creation for medical image segmentation, reconstruction, and generation. We also implement H-SeM and H-AE by employing a generic hierarchical architecture for vertical feature refinement and fusion. Extensive experiments on multiple datasets with multiple modalities showcase that this design improves model performance across different encoders.
Links & Resources
Publication
| Guoqing Zhang, Jingyun Yang and Yang Li. Flemme: A Flexible and Modular Learning Platform for Medical Images. In 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2024. | ppt |
@inproceedings{zhang2024flemme,
title = {Flemme: A Flexible and Modular Learning Platform for Medical Images},
author = {Zhang, Guoqing and Yang, Jingyun and Li, Yang},
booktitle = {2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)},
year = {2024},
publisher = {IEEE}
}